DINOSim: Zero-Shot Object Detection and Semantic Segmentation on Electron Microscopy Images
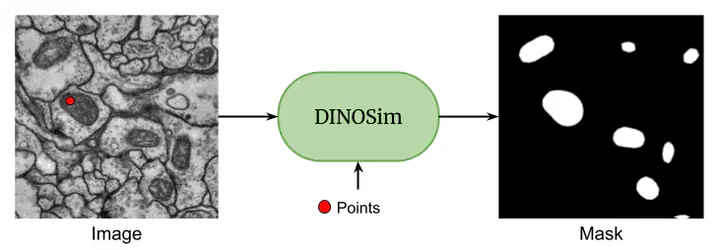 DINOSim enables zero-shot image segmentation by selecting reference points on an image.
DINOSim enables zero-shot image segmentation by selecting reference points on an image.Abstract
We present DINOSim, a novel approach leveraging the DINOv2 pretrained encoder for zero-shot object detection and segmentation in electron microscopy datasets. By exploiting semantic embeddings, DINOSim generates pseudo-labels from patch distances to a user-selected reference, which are subsequently employed in a k-nearest neighbors framework for inference. Our method effectively detects and segments previously unseen objects in electron microscopy images without additional finetuning or prompt engineering. We also investigate the impact of prompt selection and model size on accuracy and generalization. To promote accessibility, we developed an open-source Napari plugin, enabling streamlined application in scientific research. DINOSim offers a flexible and efficient solution for object detection in resource-constrained settings, addressing a critical gap in bioimage analysis.