BiaPy: Accessible deep learning on bioimages
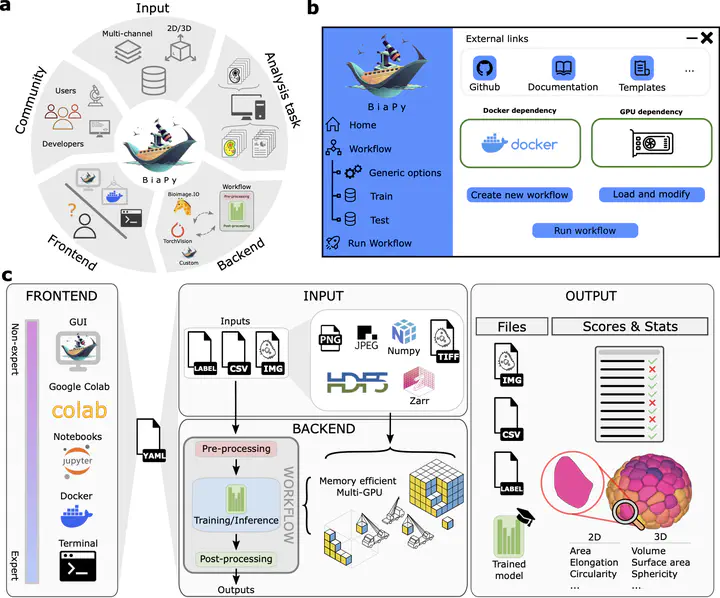 BiaPy environment and scope. BiaPy is a flexible platform for deep learning-based bioimage analysis, supporting both life science users and developers. It processes 2D/3D microscopy data through customizable workflows for tasks like segmentation, classification, and detection. Users can interact via a user-friendly GUI with a step-by-step wizard or use command-line tools. BiaPy supports multiple file formats (TIFF, H5, Zarr), multi-GPU setups, and model integration via the BioImage Model Zoo and TorchVision. Its workflows follow a three-stage structure: pre-processing, model training/inference, and post-processing, with outputs that include predictions, metrics, and analysis reports.
BiaPy environment and scope. BiaPy is a flexible platform for deep learning-based bioimage analysis, supporting both life science users and developers. It processes 2D/3D microscopy data through customizable workflows for tasks like segmentation, classification, and detection. Users can interact via a user-friendly GUI with a step-by-step wizard or use command-line tools. BiaPy supports multiple file formats (TIFF, H5, Zarr), multi-GPU setups, and model integration via the BioImage Model Zoo and TorchVision. Its workflows follow a three-stage structure: pre-processing, model training/inference, and post-processing, with outputs that include predictions, metrics, and analysis reports.Abstract
BiaPy is an open-source library and application that streamlines the use of common deep learning approaches for bioimage analysis. Designed to simplify technical complexities, it offers an intuitive interface, zero-code notebooks, and Docker integration, catering to both users and developers. While focused on deep learning workflows for 2D and 3D image data, it enhances performance with multi-GPU capabilities, memory optimization, and scalability for large datasets. Although BiaPy does not encompass all aspects of bioimage analysis, such as visualization and manual annotation tools, it empowers researchers by providing a ready-to-use environment with customizable templates that facilitate sophisticated bioimage analysis workflows.
Type
Publication
Nature Methods