New AI Tool Scans STORM Images to Distinguish Cancer Cells from Normal Cells
Our researcher Ignacio Arganda-Carreras, and PhD students Aitor González-Marfil and Daniel Franco-Barranco, collaborated in a novel deep learning method called AINU (Artificial Intelligence of the Nucleus) that can identify specific nuclear signatures at nanoscale resolution.
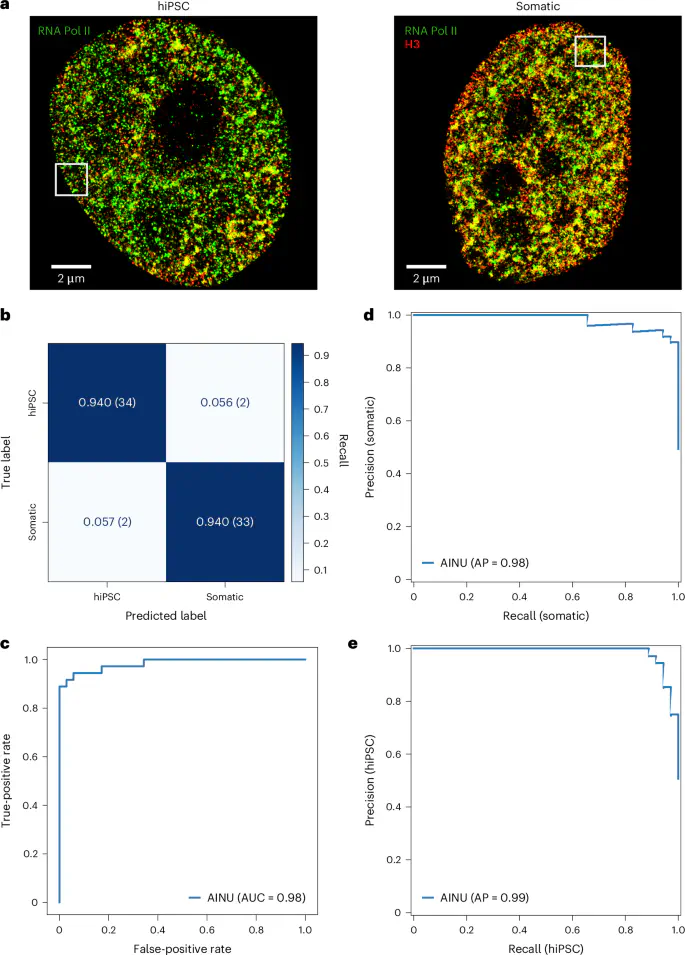
A novel deep learning method called AINU (Artificial Intelligence of the Nucleus) can identify specific nuclear signatures at nanoscale resolution. Using a small number of images as training data, AINU accurately identified human somatic cells, human induced pluripotent stem cells (hiPSCs), and differentiated cancer cells from normal cells. It also detected early-stage infected cells with DNA herpes simplex virus type 1 (HSV-1) by analyzing the spatial arrangement of core histone H3, RNA polymerase II (Pol II), or DNA from super-resolution microscopy images.
Published in Nature Machine Intelligence, this work paves the way for improved diagnostic techniques and new disease monitoring strategies. AINU, a convolutional neural network, scans high-resolution images obtained using STochastic Optical Reconstruction Microscopy (STORM), revealing structures as small as 20 nm.
The AI’s ability to detect changes in a cell’s nucleus soon after infection could help doctors monitor diseases, personalize treatments, and improve patient outcomes. Researchers believe this technology could be used in hospitals and clinics for quick and accurate diagnosis from simple blood or tissue samples.
🔗 Link to the article: https://www.nature.com/articles/s42256-024-00883-x